HRD
(Homologous Recombination Deficiency Test)
GC Genome’s HRD test provides ovarian cancer patients
with increased opportunities for targeted therapy.

What is HRD (Homologous Recombination Deficiency)?
![[GBU]홈페이지-리뉴얼_콘텐츠-이미지6-19](http://43.202.52.227/wp-content/uploads/2023/07/GBU홈페이지-리뉴얼_콘텐츠-이미지6-19.png)
When double-strand breaks occur due to DNA damage, normal cells undergo a DNA repair process called homologous recombination repair (HRR) with the assistance of various genes, including BRCA1 and BRCA2. However, cells with deficiencies in this process fail to successfully repair DNA due to issues with the assisting genes or various unidentified problems. Tumor cells that experience problems in the homologous recombination repair process are referred to as having homologous recombination deficiency (HRD).
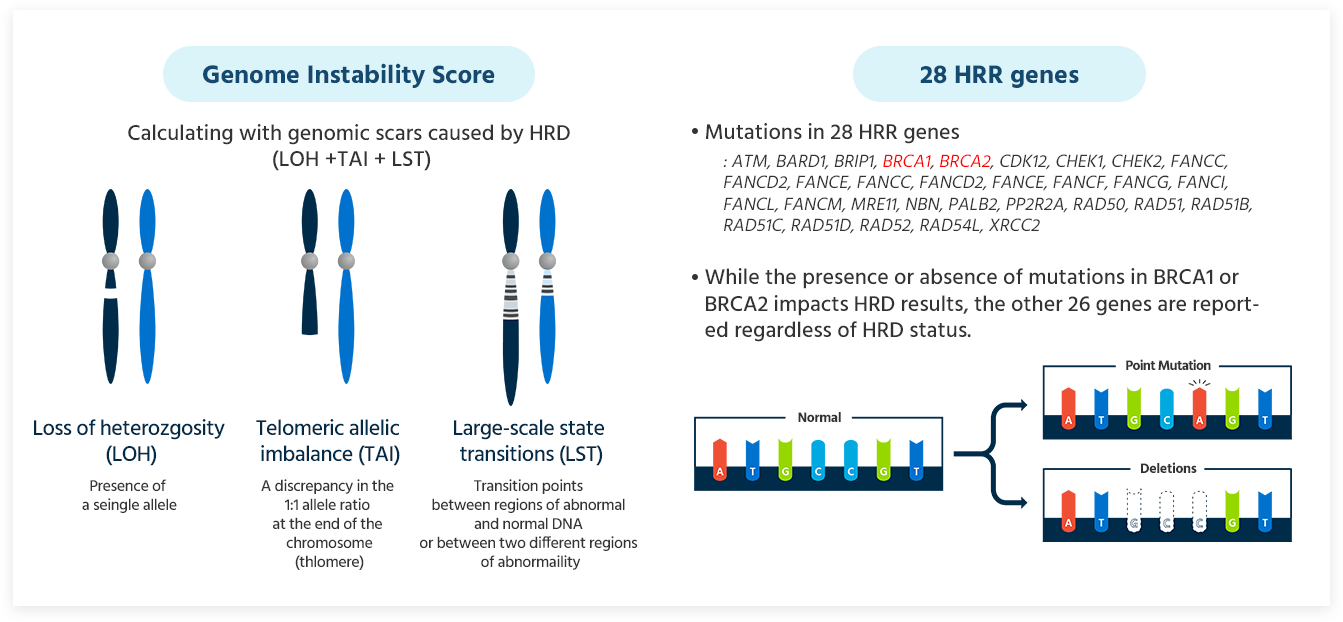
HRD testing evaluates the genomic instability by analyzing somatic mutations in the key genes BRCA1 and BRCA2, as well as three types of genomic scars generated by homologous recombination deficiency. This comprehensive analysis scores the genomic instability and assesses the HRD status.
Why is the HRD test important?
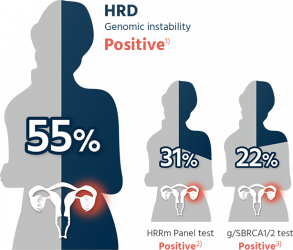
50% of the high-grade serous epithelial ovarian cancer patient is HRD positive¹
- In high-grade serous ovarian cancer patients, 50% exhibit a state of homologous recombination deficiency (HRD positive) while conventional tests may miss 20-30% of ovarian cancer patients with homologous recombination deficiency.
- Ovarian cancer has the worst prognosis of all gynecologic malignancies.
- HRD test in ovarian cancer is best assessed, not in terms of biological HRD status, but in terms of PARP inhibitor benefits.
Using PARP inhibitors has shown a benefit in progression-free survival for HRD-positive patients, both with and without BRCA mutations.
- According to the NCCN guidelines, the HRD status without g/BRCA 1/2 mutation may helpful to decide the PARP inhibitor therapy.²
- According to the ASCO guidelines, PARP inhibitor therapy is helpful to epithelial ovarian cancer patients who meet all three of the following list.³
- Patients who relapsed within 6 months of platinum-based chemotherapy
- Patients with g/sBRCA1/2 mutation or problems with Genomic instability
- Patients without experience of PARP inhibitors
![[GBU]홈페이지-리뉴얼_콘텐츠-이미지6-25](http://43.202.52.227/wp-content/uploads/2023/07/GBU홈페이지-리뉴얼_콘텐츠-이미지6-25.png)
![[GBU]홈페이지 리뉴얼_콘텐츠 이미지6-27](http://43.202.52.227/wp-content/uploads/2023/07/GBU홈페이지-리뉴얼_콘텐츠-이미지6-27.png)
Who Would Benefit from HRD Testing?
1. Newly diagnosed Ovarian cancer patients who have not undergone genetic testing
HRD testing provides a comprehensive assessment of both somatic BRCA1/BRCA2 mutations and genomic instability, essential factors for ovarian cancer patients.
2. Ovarian cancer patients with negative results in BRCA1/BRCA2 testing
HRD testing offers a greater opportunity for targeted therapy to a larger group of patients compared to hereditary and somatic BRCA1/BRCA2 testing alone.
3. Ovarian cancer patients prior to undergoing maintenance or therapeutic treatment with PARP inhibitors
Patients with positive HRD test results can expect better treatment outcomes compared to those with negative results.
Reference
1. Konstantinopoulos, Panagiotis A., et al. Cancer discovery (2015)
2. NCCN Guidelines-Ovarian Cancer including fallopian tube cancer and primary peritoneal cancer_Version 2.2020
3. Tew, William P., et al. Obstetrical & Gynecological Survey (2020)
What is HRD (Homologous Recombination Deficiency)?
When double-strand breaks occur due to DNA damage, normal cells undergo a DNA repair process called homologous recombination repair (HRR) with the assistance of various genes, including BRCA1 and BRCA2. However, cells with deficiencies in this process fail to successfully repair DNA due to issues with the assisting genes or various unidentified problems. Tumor cells that experience problems in the homologous recombination repair process are referred to as having homologous recombination deficiency (HRD).
HRD testing evaluates the genomic instability by analyzing somatic mutations in the key genes BRCA1 and BRCA2, as well as three types of genomic scars generated by homologous recombination deficiency. This comprehensive analysis scores the genomic instability and assesses the HRD status.
![[GBU]홈페이지-리뉴얼_콘텐츠-이미지6-17](http://43.202.52.227/wp-content/uploads/2023/07/GBU홈페이지-리뉴얼_콘텐츠-이미지6-17.png)
![[GBU]홈페이지-리뉴얼_콘텐츠-이미지6-19](http://43.202.52.227/wp-content/uploads/2023/07/GBU홈페이지-리뉴얼_콘텐츠-이미지6-19.png)
50% of the high-grade serous epithelial ovarian cancer patient is HRD positive
- In high-grade serous ovarian cancer patients, 50% exhibit a state of homologous recombination deficiency (HRD positive) while conventional tests may miss 20-30% of ovarian cancer patients with homologous recombination deficiency.
- Ovarian cancer has the worst prognosis of all gynecologic malignancies.
- HRD test in ovarian cancer is best assessed, not in terms of biological HRD status, but in terms of PARP inhibitor benefits.
HRD: Homologous Recombination Deficiency, HRRm: Homologous Recombination Repair mutation,
gBRCA1/2: germline BRCA1/2, sBRCA1/2: somatic BRCA1/2
1. Ray-Coquard, Isabelle, et al. New England Journal of Medicine 381.25 (2019)
2. Norquist, B. S., et al. Gynecologic Oncology 141 (2016)
3. Yates, Melinda, et al. Annals of Oncology 25
Using PARP inhibitors has shown a benefit in progression-free survival for HRD-positive patients, both with and without BRCA mutations.
- Patients who relapsed within 6 months of platinum-based chemotherapy
- Patients with g/sBRCA1/2 mutation or problems with Genomic instability
- Patients without experience of PARP inhibitors
GCG HRD Test provides a comprehensive result that includes the Genomic Instability Score (GIS) and HRR genes including BRCA1 and BRCA2.
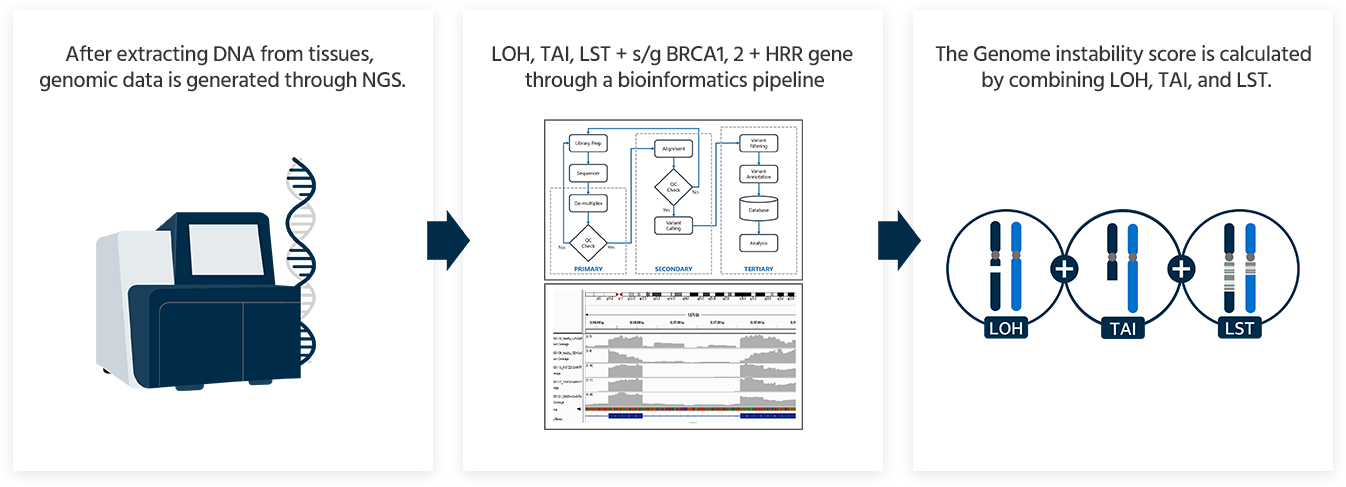
Genomic Instability Score(GIS)

28 HRR genes
- Mutations in 28 HRR genes:
*ATM, BARD1, BRIP1, BRCA1, BRCA2, CDK12, CHEK1, CHEK2, FANCC, FANCD2, FANCE, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCL, FANCM, MRE11, NBN, PALB2, PP2R2A, RAD50, RAD51, RAD51B, RAD51C, RAD51D, RAD52, RAD54L, XRCC2* - While the presence or absence of mutations in BRCA1 or BRCA2 impacts HRD results, the other 26 genes are reported regardless of HRD status.
HRD Test is based on GIS 42 scores.
Providing a positive/negative test result based on GIS 42 scores used in clinical trials.
Table 1. Univarate Cox PH analysis of PFS and OS for HRD and tBRCA
| Variable | Levels | Overall cohort | HGSOC supset | ||
|---|---|---|---|---|---|
| HR(95% CI) | P | HR (95% CI) | P | ||
| PFS | |||||
| HRD status (≥42) | HRD positive | 0.65 (0.46-0.93) | 0.014 | 0.50 (0.34-0.73) | 0.00021 |
| HRD negative | Ref | Ref | |||
| HRD status (≥33) | HRD positive | ND | ND | 0.51 (0.36-0.72) | 0.00014 |
| HRD negative | ND | Ref | |||
| tBRCA mutation status | Mutant | 0.61 (0.38-0.99) | 0.034 | 0.48 (0.29-0.79) | 0.0017 |
| Wild-type | Ref | Ref | |||
| CCNE1 amplification statusa | Amplified | 1.91 (1.32-2.75) | 0.0011 | 1.56 (1.04-2.34) | 0.038 |
| Not amplified | Ref | Ref | |||
| OS | |||||
| HRD status (≥42) | HRD positive | 0.57 (0.36-0.92) | 0.016 | 0.45 (0.27-0.74) | 0.0011 |
| HRD negative | Ref | Ref | |||
| HRD status (≥33) | HRD positive | ND | ND | 0.43 (0.27-0.69) | 0.00033 |
| HRD negative | ND | Ref | |||
| tBRCA mutation status | Mutant | 0.64 (0.35-1.17) | 0.12 | 0.50 (0.26-0.95) | 0.022 |
| Wild-type | Ref | Ref | |||
| CCNE1 amplification statusa | Amplified | 1.82 (1.15-2.88) | 0.015 | 1.72 (1.04-2.85) | 0.043 |
| Not amplified | Ref | Ref | |||
aCCNE1 amplification status was determined fot 248 of 250 patients in the full cohort, and 178 of 179 patients in the HGSOC subcohort.
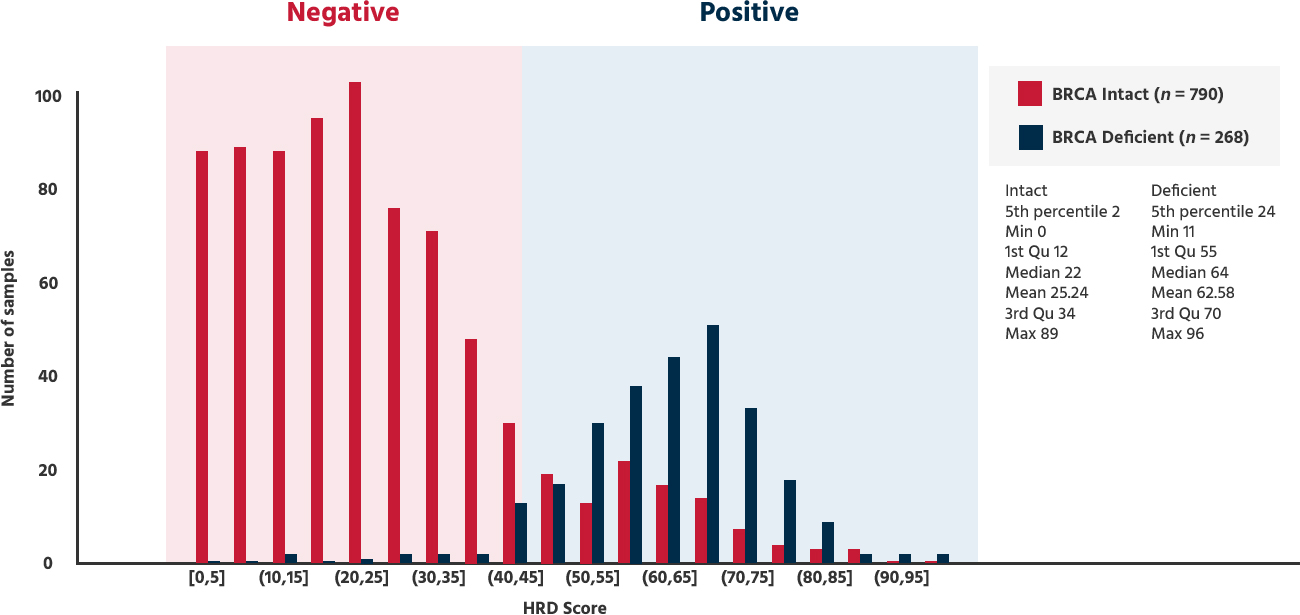
If the GIS(Genome instability score) is 42 or higher, or if BRCA is positive → HRD positive
| GIS ≥ 42 | HRD Positive → PAPRi Sensitive |
| GIS < 42 | HRD Negative → PAPRi Less Sensitive |
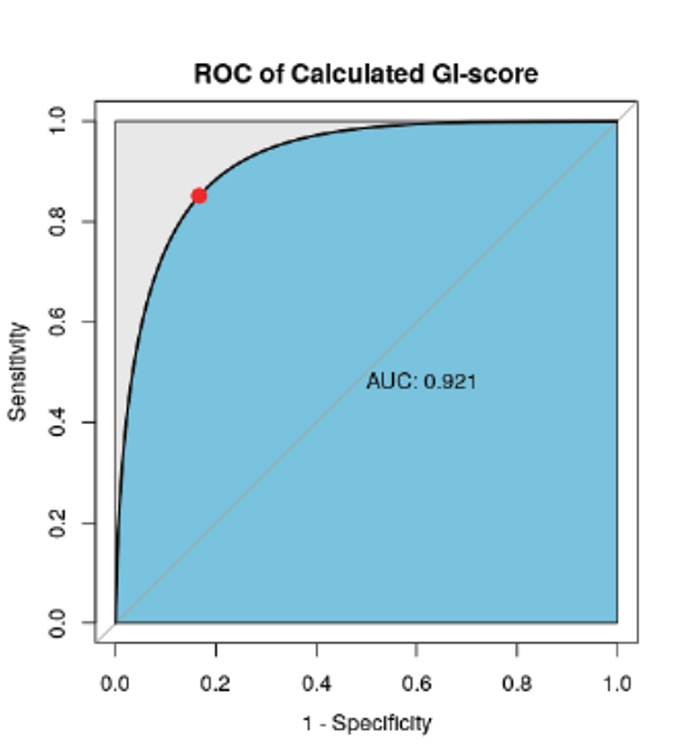
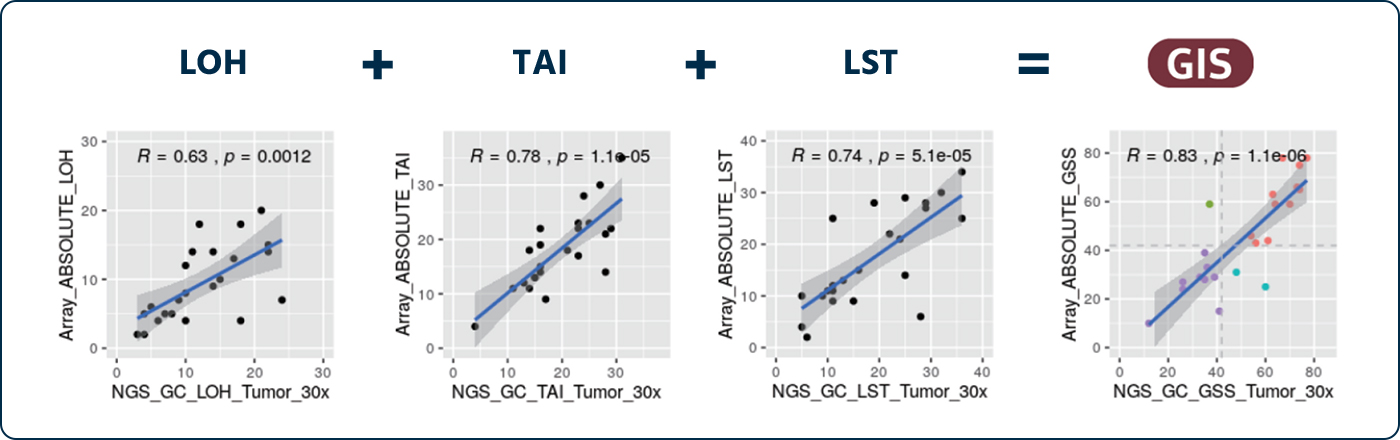
Performance Evaluation of Methods and Algorithms by Comparison of HRD Score.
HRD(NGS) vs SNP-Array (R = 0.83)
By comparing the GC Genome’s HRD test method(NGS) with SNP array method in 26 samples of ovarian paents reported by TCGA/ICGC, it is confirmed that both test methods are equivalent.
HRD vs HRDetect(public data) (AUC = 0.921)
- “HRDetect” reported in the paper is a test that calcuting HRD score with the algorithm, which is combinating LOH and multiple mutational signatures.
- By comparing the result data from GC Genome’s algorithm with HRDetect’s algorithm in 50 ovarian cancer patients reported as public data, it is confirmed that the performance of two test algorithms are equivalent. Reference: Nat Med. 2017 Apr;23(4):517-525